Protein Blast Analysis . Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. Blastp simply compares a protein query to a protein database. This blast quickstart chapter illustrates the use of the principal blast programs to solve problems that arise in the analysis of protein. “blast” (basic local alignment search. Compares one or more protein query sequences to a subject protein sequence or a database of protein. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. To build upon these protein data and to aid analysis, uniprot provides four main tools: Blast stands for basic local alignment search tool.
from proteopedia.org
The basic local alignment search tool (blast) finds regions of local similarity between sequences. “blast” (basic local alignment search. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. This blast quickstart chapter illustrates the use of the principal blast programs to solve problems that arise in the analysis of protein. Blast stands for basic local alignment search tool. Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. Blastp simply compares a protein query to a protein database. To build upon these protein data and to aid analysis, uniprot provides four main tools: Compares one or more protein query sequences to a subject protein sequence or a database of protein.
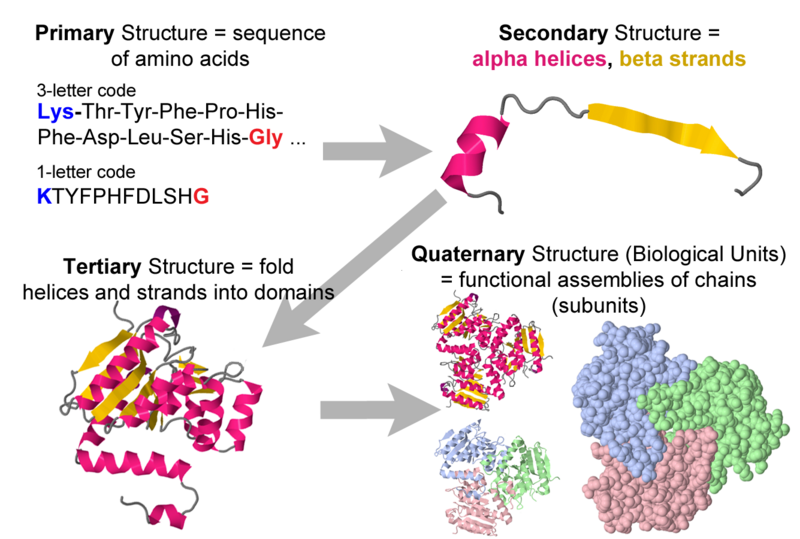
Four levels of protein structure Proteopedia, life in 3D
Protein Blast Analysis Blast stands for basic local alignment search tool. This blast quickstart chapter illustrates the use of the principal blast programs to solve problems that arise in the analysis of protein. Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. Blastp simply compares a protein query to a protein database. “blast” (basic local alignment search. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Compares one or more protein query sequences to a subject protein sequence or a database of protein. To build upon these protein data and to aid analysis, uniprot provides four main tools: Blast stands for basic local alignment search tool.
From www.researchgate.net
(PDF) Beyond BLASTing Tertiary and Quaternary Structure Analysis Helps Protein Blast Analysis Compares one or more protein query sequences to a subject protein sequence or a database of protein. Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp simply compares a protein. Protein Blast Analysis.
From www.manuscriptedit.com
The future of protein analysis Beyond traditional BLAST? Protein Blast Analysis Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. Compares one or more protein query sequences to a subject protein sequence or a database of protein. The basic local alignment search tool (blast) finds regions of local similarity between sequences. This blast quickstart chapter illustrates. Protein Blast Analysis.
From www.researchgate.net
Blasting the sequences of Vipp proteins in different species. (A Protein Blast Analysis Compares one or more protein query sequences to a subject protein sequence or a database of protein. Blast stands for basic local alignment search tool. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. “blast” (basic local alignment search. Today, one of the most common tools. Protein Blast Analysis.
From www.creative-proteomics.com
Bioinformatics for Protein Creative Proteomics Protein Blast Analysis Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. This blast quickstart chapter illustrates the use of the principal blast programs to solve problems that arise in the analysis of protein. Blast stands for basic local alignment search tool. Compares one or more protein query. Protein Blast Analysis.
From sgbc.github.io
Blast Online Bioinformatics course Protein Blast Analysis The basic local alignment search tool (blast) finds regions of local similarity between sequences. “blast” (basic local alignment search. Blast stands for basic local alignment search tool. To build upon these protein data and to aid analysis, uniprot provides four main tools: A blast search enables a researcher to compare a subject protein (called a query) with a database of. Protein Blast Analysis.
From www.youtube.com
BLAST Protein YouTube Protein Blast Analysis Compares one or more protein query sequences to a subject protein sequence or a database of protein. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. Blast stands for basic local alignment search tool. Blastp simply compares a protein query to a protein database. Today, one. Protein Blast Analysis.
From abacus.bates.edu
BLASTP Results Part 1 Protein Blast Analysis To build upon these protein data and to aid analysis, uniprot provides four main tools: This blast quickstart chapter illustrates the use of the principal blast programs to solve problems that arise in the analysis of protein. Blast stands for basic local alignment search tool. A blast search enables a researcher to compare a subject protein (called a query) with. Protein Blast Analysis.
From www.slideserve.com
PPT Protein Analysis (Beyond BLAST) PowerPoint Presentation, free Protein Blast Analysis Compares one or more protein query sequences to a subject protein sequence or a database of protein. Blast stands for basic local alignment search tool. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. This blast quickstart chapter illustrates the use of the principal blast programs. Protein Blast Analysis.
From www.researchgate.net
Blast analysis results of BrRGF6 candidate interacting protein Protein Blast Analysis Blast stands for basic local alignment search tool. Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. Blastp simply compares a protein query to a protein database. To build upon these protein data and to aid analysis, uniprot provides four main tools: “blast” (basic local. Protein Blast Analysis.
From usuhs.libguides.com
BLASTP Protein Sequence Search Introduction to NCBI Bioinformatics Protein Blast Analysis To build upon these protein data and to aid analysis, uniprot provides four main tools: A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. Blastp simply compares a protein query to a protein database. This blast quickstart chapter illustrates the use of the principal blast programs. Protein Blast Analysis.
From usuhs.libguides.com
BLAST Results Introduction to NCBI Bioinformatics Resources Protein Blast Analysis “blast” (basic local alignment search. Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. Blast stands for basic local alignment search tool.. Protein Blast Analysis.
From usuhs.libguides.com
BLASTP Protein Sequence Search Introduction to NCBI Bioinformatics Protein Blast Analysis To build upon these protein data and to aid analysis, uniprot provides four main tools: Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Blastp simply compares a protein query to. Protein Blast Analysis.
From www.youtube.com
NCBI BLAST tutorial how to use blast for finding and aligning DNA or Protein Blast Analysis This blast quickstart chapter illustrates the use of the principal blast programs to solve problems that arise in the analysis of protein. Blast stands for basic local alignment search tool. “blast” (basic local alignment search. To build upon these protein data and to aid analysis, uniprot provides four main tools: A blast search enables a researcher to compare a subject. Protein Blast Analysis.
From www.youtube.com
Which BLAST Algorithm to use for ProteinProtein BLAST YouTube Protein Blast Analysis Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. Blast stands for basic local alignment search tool. The basic local alignment search tool (blast) finds regions of local similarity between sequences. To build upon these protein data and to aid analysis, uniprot provides four main. Protein Blast Analysis.
From www.researchgate.net
Gene clusters for secreted proteins. Left, schematic representation Protein Blast Analysis Compares one or more protein query sequences to a subject protein sequence or a database of protein. The basic local alignment search tool (blast) finds regions of local similarity between sequences. Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. “blast” (basic local alignment search.. Protein Blast Analysis.
From www.researchgate.net
Multiple protein sequence alignment of HTT exon 29. Multiple protein Protein Blast Analysis Today, one of the most common tools used to examine dna and protein sequences is the basic local alignment search tool, also known as. “blast” (basic local alignment search. The basic local alignment search tool (blast) finds regions of local similarity between sequences. This blast quickstart chapter illustrates the use of the principal blast programs to solve problems that arise. Protein Blast Analysis.
From www.slideserve.com
PPT BLAST PowerPoint Presentation, free download ID5789334 Protein Blast Analysis This blast quickstart chapter illustrates the use of the principal blast programs to solve problems that arise in the analysis of protein. To build upon these protein data and to aid analysis, uniprot provides four main tools: Blastp simply compares a protein query to a protein database. A blast search enables a researcher to compare a subject protein (called a. Protein Blast Analysis.
From www.researchgate.net
relationship and architecture of conserved protein motifs Protein Blast Analysis To build upon these protein data and to aid analysis, uniprot provides four main tools: “blast” (basic local alignment search. A blast search enables a researcher to compare a subject protein (called a query) with a database of sequences, and identify database sequences. Blast stands for basic local alignment search tool. This blast quickstart chapter illustrates the use of the. Protein Blast Analysis.